Research
Research projects:
"Chemical ecology of cross-kingdom interactions"
The focus of our research group is to elucidate the mutualistic interactions between prokaryotes and eukaryotes ("cross-kingdom cross-talk"). Our previous work has shown that challenges of chemical ecology - at the interface between biology and chemistry - can only be resolved successfully by an interdisciplinary research approach. The consistent combination of classical bioassays and chemical analytical methods reveals novel insights into the complex networks of infochemicals and signal molecules, along with their ecophysiological significance. Our research approach focuses on the fundamental understanding of the functioning of infochemicals in biocoenoses that can be mimicked in laboratory studies, after which field experiments are conducted to prove their ecological significance.
To achieve our research goals, we have established the marine green seaweed Ulva (Chlorophyta) as a powerful model organism. Our research focuses on four major areas:
I. Ulva as model organism: Symbiotic Interactions Between Bacteria and Marine Macroalgae
- Deciphering the chemical communication (“chemosphere”) between Ulva and its associated bacteria
- Understanding cross-kingdom signaling mediated by infochemicals
II. The Ulva Holobiont Under Stress
- Investigating the impact of micropollutants on Ulva and its microbiome
- Exploring how the microbiome contributes to stress resilience in Ulva
III. Regulation of Gametogenesis and Gamete Release
- Identifying sporulation inhibitors that control the differentiation of blade cells into gametangia
- Uncovering epigenetic mechanisms shaping the algal life cycle
IV. Unlocking Metal Acquisition in Microbial Communities and their Host Organism
- Identifying metallophores that facilitate metal uptake in bacteria and algae
- Measuring the bioavailability of trace metals essential for nitrogenase activity
I. Symbiotic interactions of the marine macroalgae Ulva and its associated bacteria
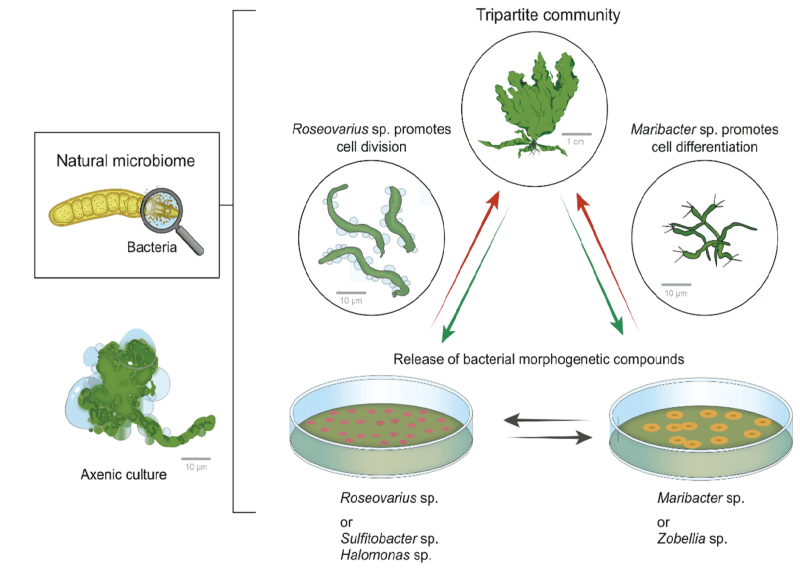
Tripartite community of Ulva and its associated bacteria.
Graphic: Thomas WichardTripartite community and representative morphotypes. Tripartite community of U. mutabilis with proposed essential interactions for standardized experimental set-ups. A combination of Roseovarius sp. (MS2) and Maribacter sp. (MS6) excreting morphogenetic substances recover growth and morphogenesis of the wildtype (shown in the top) and the mutant slender of U. mutabilis: Roseovarius sp. promotes cell division (A: scale bars = 1 mm) and Maribacter sp. promotes rhizoid and cell wall formation (B: scale bars = 1 mm). Strictly axenic cultures develop into a callus like morphotype consisting of undifferentiated cells without normal cell walls. Image: © Wichard, T. (2023) From model organism to application: Bacteria-induced growth and development of the green seaweed Ulva and the potential of microbe leveraging in algal aquaculture. Seminars in Cell & Developmental Biology 134:69-78. https://doi.org/10.1016/j.semcdb.2022.04.007External link. This project is funded by the Colaborative Research Center "Chemical Mediators In Complex Biosystems" ChemBioSys SFB 1127 - Project A01External link.
Running Project
Our recent project aims to unravel the signal-mediated cross-kingdom interactions between the marine green alga Ulva mutabilis and its associated bacteria. Morphogens such as thallusin are released by bacteria and induce diverse algal developmental processes. Thallusin derivatives will be synthesized to investigate their quantitative structure–activity relationships and to localize thallusin within Ulva using imaging techniques. Key genes and metabolites involved in thallusin homeostasis will be identified through comparative transcriptomic and metabolomic analyses. A further focus will be on the role of thallusin in commercially relevant algal aquaculture systems.
II. Ulva holobiont under stress (in progress)
III. Regulation of gametogenesis and gametes release in Ulva
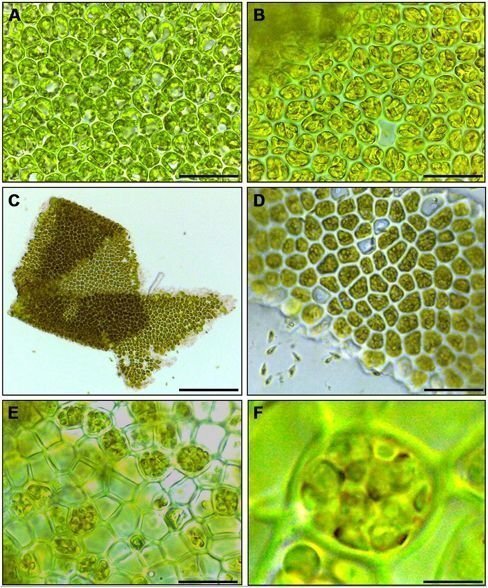
Gametogenesis
Image: Vesty et al. (2015) Frontiers in Plant ScienceInduction and regulation of gametogenesis and zoosporogenesis in Ulva mutabilis and Ulva linza. Phenotypic changes of blade cells during gametogenesis and gamete release. (A) Blade cells 24 h after induction resemble uninduced blade cells: cells are square and often in transverse rows. (B) 48 h after induction, blade cells differentiate into gametangia containing finally 16 progametes, which mature during the following night into fully developed gametes ready for swarming. (C,D) Gametes are discharged in the morning of the third day. (E) Discharged sporangia and (F) zoospores within a sporangium are shown. Gametophytes (A-D) and sporophytes (E,F) were grown under standard conditions (Scale bars: A,B,D = 25 μm; C = 140 μm, E = 16 μm, F = 4 μm). Image: © Vesty EF, Kessler RW, Wichard T and Coates JC (2015) Regulation of gametogenesis and zoosporogenesis in Ulva linza (Chlorophyta): comparison with Ulva mutabilis and potential for laboratory culture. Front. Plant Sci. 6:15. doi: 10.3389/fpls.2015.00015External link.
IV. Determination and quantification of metallophores
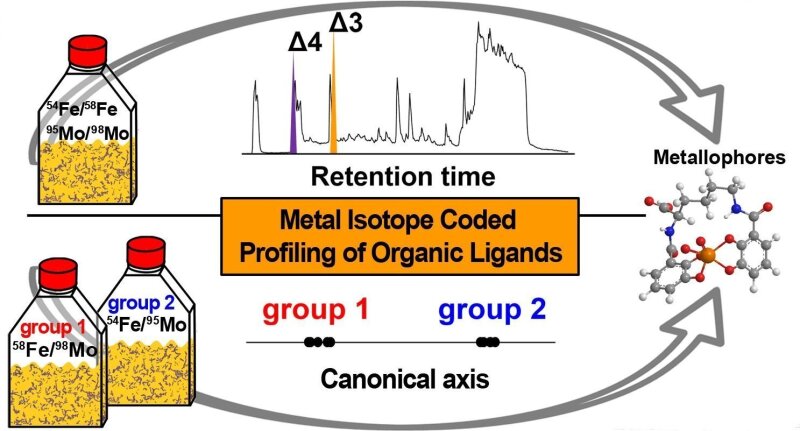
Metallophore mapping
Image: Deicke et al. (2014) AnalystMetal isotope coded profiling (MICP) introduces a universal discovery platform for metal chelating natural products that act as metallophores, ion buffers or sequestering agents. The detection of cation and oxoanion complexing ligands is facilitated by the identification of unique isotopic signatures created by the application of isotopically pure metals. Figure © Deicke, M., Mohr, J. F., Bellenger, J. P., Wichard, T. (2014) Metallophore mapping in complex matrices by metal isotope coded profiling of organic ligands. Analyst, 23, 6096 - 6099.External link
Running project
Paratuberculosis is a progressive, fatal enteritis in ruminants caused by Mycobacterium avium ssp. paratuberculosis (MAP), which primarily infects cells of the distal ileum—the site of host zinc uptake. MAP carries the canonical ZnuABC zinc transporter, as well as two additional, MAP-specific zinc importers on LSP14 and LSP15. Notably, LSP14 contains a unique zincophore gene cluster (sid), suggesting an additional zinc acquisition system not found in related mycobacteria. We previously showed that sid is zinc-regulated by Zur and encodes an NRPS potentially producing a zincophore. Using metal isotope–coded profiling (MICP), we have detected a MAP zincophore candidate for the first time.
This project is conducted in close collaboration with Dr. Elke Goethe (TiHo Hannover)External link.
It aims to identify and characterize MAP zincophores, link the sid cluster to zincophore production, determine the role of sid in zinc homeostasis and intracellular survival, and investigate zincophore translocation. The expected outcomes will advance our understanding of zinc regulation and virulence mechanisms in MAP and related mycobacteria.